Artikeln är enligt undertecknad en ren hyllning till HoloMonitor och vad PHI,s excellenta teknik förmår leverera info/kunskapsmässigt.
Jag rekommenderar alla phi,are att läsa hela rapporten. Men här kommer ett utdrag för den slöe.
Exploiting the potential of commercial digital holographic microscopy by combining it with 3D matrix cell culture assays
Published: 07 September 2020 Länk : https://www.nature.com/articles/s41598-020-71538-1#Sec8
Abstract
3D
cell culture assays are becoming increasingly popular due to their
higher resemblance to tissue environment. These provide an increased
complexity compared to the growth on 2D surface and therefore allow
studies of advanced cellular properties such as invasion. We report here
on the use of 3D Matrigel cell preparations combined with a particular
gentle and informative type of live-cell microscopy: quantitative
digital holographic microscopy (DHM), here performed by a commercial
software-integrated system, currently mostly used for 2D cell culture
preparations. By demonstrating this compatibility, we highlight the
possible time-efficient quantitative analysis obtained by using a
commercial software-integrated DHM system, also for cells in a more
advanced 3D culture environment. Further, we demonstrate two very
different examples making use of this advantage by performing
quantitative DHM analysis of: (1) wound closure cell monolayer Matrigel
invasion assay and (2) Matrigel-trapped single and clumps of suspension
cells. For both these, we benefited from the autofocus functionality of
digital phase holographic imaging to obtain 3D information for cells
migrating in a 3D environment. For the latter, we demonstrate that it is
possible to quantitatively measure tumourigenic properties like growth
of cell clump (or spheroid) over time, as well as single-cell invasion
out of cell clump and into the surrounding extracellular matrix.
Overall, our findings highlight several possibilities for 3D digital
holographic microscopy applications combined with 3D cell preparations,
therein studies of drug response or genetic alterations on invasion
capacity as well as on tumour growth and metastasis.
Matrigel is one example of a commonly used reagent that mimics the basement membrane and contains main components of several extracellular matrix (ECM) structural proteins, including collagen IV and laminin. Matrigel forms a gel-like matrix, thereby providing a more complex extracellular environment. In such a three-dimensional landscape, it may be possible for cells to adapt a more in vivo-like migratory mode in which they digest, attach to and navigate through pores of the matrix by proteolysis, integrin expression and intracellular contraction, as well as form invading structures. The classical migration assay, in which a scratch wound is made in a confluent cell culture followed by monitoring of the two cell fronts rejoining to close the gap, may be performed in Matrigel as a model for invasion and these types of models are popular in cancer studies, such as for drug discovery.
The most typical microscopy technique used for these types of wound-healing migration or invasion assays, is regular phase contrast and differential interference contrast (DIC) microscopy. An alternative label-free live-cell imaging technique that additionally provides cell 3D information is digital holographic microscopy (DHM), a type of quantitative phase imaging (QPI).
The present work focuses on the use of DHM in imaging cell culture populations, which is a recognized accompaniment to higher-magnification super-resolution imaging of subcellular structures. Due to the low light intensity of the laser light used in QPI methods like DHM, live-cell imaging can be performed with frequent image acquisition and is suitable for long-term monitoring of single cells or cell population morphology. This microscope technique records a digital hologram of the cell, and this recorded interference pattern is processed computationally to produce a quantitative phase shift image, a holographic image. The digital reconstruction of the hologram is performed numerically in DHM. Therefore, these digital images contain information on biophysical parameters, such as cell thickness, volume and shape, which can be quantified and used to monitor cell morphology in for example phenotype or drug screens.
The last decades, QPI has emerged as an important method in biomedical imaging and various QPI techniques have been developed and implemented as tools for biomedical research. This imaging technology is developing towards a more application-based field and commercially available QPI systems have recently been made available. In the present study, we used a commercial holographic microscope with integrated software for various applications for quantitative analysis of several cell morphological features in addition to single-cell tracking and wound analysis (see “Methods” section for details).
DHM live-cell imaging is indeed applied in migration studies, both single-cell tracking as well as collective wound migration. The latter allow for visualization of the migrating cell layer and quantitative motility data based on cell-covered area. A study using digital holographic microscopy provided additional detailed information on morphological features extracted from this type of assay, like cell layer thickness and identification of proliferating single cells from the holographic images. Another study using this system, showed that it was well suited for monitoring and quantitatively assess the motile capacity of cells migrating in a wound-healing assay compared to other established migration assay, like transwell assays. In addition, these authors concluded that this commercial all-in-one DHM system provided advantages such as reproducibility of measurements and compatibility with high throughput applications.
Although DHM is extensively used and optimized for wound-healing migration analysis, the potential of live-cell DHM imaging has, as far as we know, not been extensively explored for invasion studies using dense gel matrices, like thick Matrigel preparations, covering the wound gap. Interference caused by light scattering effects during imaging of such preparations has been indicated as a challenge for DHM imaging, albeit preparations with collagen did prove compatible as well as a Matrigel cluster assay on fixed cells. Cells chemotactically migrating in diluted Matrigel were also trackable using DHM. To our knowledge, there are at present limited scientific literature reporting on the performance of the newest commercial DHM imaging technology for wound analysis of cells embedded in thick Matrigel.
The aim of this report was to evaluate the compatibility between a commercial all-in-one DHM imaging system and cell cultures embedded in 3D Matrigel matrix. Hence, we wanted to test whether it would be possible to achieve a double benefit from the strength of monolayer 3D cell preparations and the DHM imaging and its accompanying software application for easy and robust quantifications of cells embedded in and invading into Matrigel.
By further taking advantage of the compatibility of this DHM system with Matrigel, we performed analyses of matrix-immobilized suspension cells and were able to measure two interesting features connected to tumour biology: (1) growth of cell clump over time, and (2) single-cell invasion out of cell clump and into the surrounding Matrigel matrix.
Some limitations of the DHM system should be noted. As a cell population-based instrument with a 20 × objective, this DHM system has limitations regarding detection and quantification of fine protrusions as possible with other systems. Nor is it possible to distinguish intracellular organelles (besides the nucleus to some degree), something which has been done with other QPI systems. Additionally, the commercial system used here is suitable for high-throughput data analyses and is developed for adhesive cells in monolayers. We have shown here that it is possible to challenge this system past adhesive monolayers with a good performance analyzing cells in 3D Matrigel in all applications tested. Nevertheless, there is a limit as cells or cell clumps must not exceed a certain size range. It should also be noted that the experiments performed here were all on cell monolayers embedded in 3D matrix. A true tracking of cells in a full 3D environment in which cell migration can also be followed in z- in addition to x/y-plane, is not possible with the DHM system reported on here.
The strong benefits of the commercial DHM system used here, is the all-in-one integration with a user-friendly accompanying software with a multitude of analysis options and quantifiable cell parameters.
3D cell culture advancements are becoming increasingly popular in all aspects of cell biological research. Our findings expand the use of digital holographic microscopy (DHM) designed for monolayer adhesive cells to reveal novel characteristics of cancer cell invasion in 3D and highlight its potential to contribute to our understanding of invasive mechanisms through for example large-scale studies of drug response on invasion capacity as well as on tumour growth and metastasis.
Methods
Introduction
It is increasingly recognized that 3D cell culture assays mimic the physiological condition with higher fidelity compared to 2D substrates. In particular in cancer research, various in vitro systems have been developed to enable the study of cancer cell biology in a 3D context. Motility assays, such as in vitro wound healing assays are widely applied to study directional migratory capacity of cancer cells. With the addition of a 3D extracellular matrix, one can more closely mimic the physiological biomechanics of various cellular processes involved in migration that influence on cancer cell invasion mode and the metastatic dissemination of primary cancers. In vitro-formed matrix gels is a good cell culture model for cancer cell invasion. It is often used in combination with multicellular spheroids, measuring tumour growth and sprouting.Matrigel is one example of a commonly used reagent that mimics the basement membrane and contains main components of several extracellular matrix (ECM) structural proteins, including collagen IV and laminin. Matrigel forms a gel-like matrix, thereby providing a more complex extracellular environment. In such a three-dimensional landscape, it may be possible for cells to adapt a more in vivo-like migratory mode in which they digest, attach to and navigate through pores of the matrix by proteolysis, integrin expression and intracellular contraction, as well as form invading structures. The classical migration assay, in which a scratch wound is made in a confluent cell culture followed by monitoring of the two cell fronts rejoining to close the gap, may be performed in Matrigel as a model for invasion and these types of models are popular in cancer studies, such as for drug discovery.
The most typical microscopy technique used for these types of wound-healing migration or invasion assays, is regular phase contrast and differential interference contrast (DIC) microscopy. An alternative label-free live-cell imaging technique that additionally provides cell 3D information is digital holographic microscopy (DHM), a type of quantitative phase imaging (QPI).
The present work focuses on the use of DHM in imaging cell culture populations, which is a recognized accompaniment to higher-magnification super-resolution imaging of subcellular structures. Due to the low light intensity of the laser light used in QPI methods like DHM, live-cell imaging can be performed with frequent image acquisition and is suitable for long-term monitoring of single cells or cell population morphology. This microscope technique records a digital hologram of the cell, and this recorded interference pattern is processed computationally to produce a quantitative phase shift image, a holographic image. The digital reconstruction of the hologram is performed numerically in DHM. Therefore, these digital images contain information on biophysical parameters, such as cell thickness, volume and shape, which can be quantified and used to monitor cell morphology in for example phenotype or drug screens.
The last decades, QPI has emerged as an important method in biomedical imaging and various QPI techniques have been developed and implemented as tools for biomedical research. This imaging technology is developing towards a more application-based field and commercially available QPI systems have recently been made available. In the present study, we used a commercial holographic microscope with integrated software for various applications for quantitative analysis of several cell morphological features in addition to single-cell tracking and wound analysis (see “Methods” section for details).
DHM live-cell imaging is indeed applied in migration studies, both single-cell tracking as well as collective wound migration. The latter allow for visualization of the migrating cell layer and quantitative motility data based on cell-covered area. A study using digital holographic microscopy provided additional detailed information on morphological features extracted from this type of assay, like cell layer thickness and identification of proliferating single cells from the holographic images. Another study using this system, showed that it was well suited for monitoring and quantitatively assess the motile capacity of cells migrating in a wound-healing assay compared to other established migration assay, like transwell assays. In addition, these authors concluded that this commercial all-in-one DHM system provided advantages such as reproducibility of measurements and compatibility with high throughput applications.
Although DHM is extensively used and optimized for wound-healing migration analysis, the potential of live-cell DHM imaging has, as far as we know, not been extensively explored for invasion studies using dense gel matrices, like thick Matrigel preparations, covering the wound gap. Interference caused by light scattering effects during imaging of such preparations has been indicated as a challenge for DHM imaging, albeit preparations with collagen did prove compatible as well as a Matrigel cluster assay on fixed cells. Cells chemotactically migrating in diluted Matrigel were also trackable using DHM. To our knowledge, there are at present limited scientific literature reporting on the performance of the newest commercial DHM imaging technology for wound analysis of cells embedded in thick Matrigel.
The aim of this report was to evaluate the compatibility between a commercial all-in-one DHM imaging system and cell cultures embedded in 3D Matrigel matrix. Hence, we wanted to test whether it would be possible to achieve a double benefit from the strength of monolayer 3D cell preparations and the DHM imaging and its accompanying software application for easy and robust quantifications of cells embedded in and invading into Matrigel.
Conclusion and future perspectives
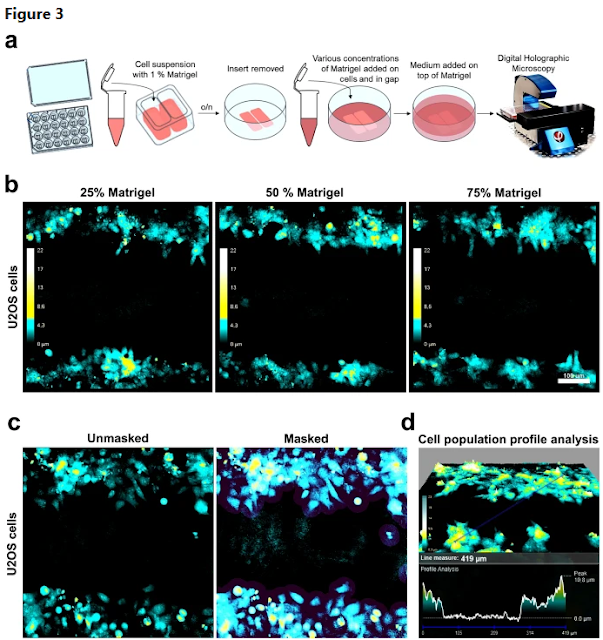
We here demonstrated the compatibility of Matrigel preparations with a commercial software-integrated system for digital holographic microscopy and quantitative analysis. Neither image quality nor quantitative analysis was notably affected, even by thick Matrigel preparations (50–75%). We took advantage of this and challenged the integrated wound-healing analysis tool that is normally used for migration studies, with analysis of invasion capacity in Matrigel-covered cell gap.By further taking advantage of the compatibility of this DHM system with Matrigel, we performed analyses of matrix-immobilized suspension cells and were able to measure two interesting features connected to tumour biology: (1) growth of cell clump over time, and (2) single-cell invasion out of cell clump and into the surrounding Matrigel matrix.
Some limitations of the DHM system should be noted. As a cell population-based instrument with a 20 × objective, this DHM system has limitations regarding detection and quantification of fine protrusions as possible with other systems. Nor is it possible to distinguish intracellular organelles (besides the nucleus to some degree), something which has been done with other QPI systems. Additionally, the commercial system used here is suitable for high-throughput data analyses and is developed for adhesive cells in monolayers. We have shown here that it is possible to challenge this system past adhesive monolayers with a good performance analyzing cells in 3D Matrigel in all applications tested. Nevertheless, there is a limit as cells or cell clumps must not exceed a certain size range. It should also be noted that the experiments performed here were all on cell monolayers embedded in 3D matrix. A true tracking of cells in a full 3D environment in which cell migration can also be followed in z- in addition to x/y-plane, is not possible with the DHM system reported on here.
The strong benefits of the commercial DHM system used here, is the all-in-one integration with a user-friendly accompanying software with a multitude of analysis options and quantifiable cell parameters.
3D cell culture advancements are becoming increasingly popular in all aspects of cell biological research. Our findings expand the use of digital holographic microscopy (DHM) designed for monolayer adhesive cells to reveal novel characteristics of cancer cell invasion in 3D and highlight its potential to contribute to our understanding of invasive mechanisms through for example large-scale studies of drug response on invasion capacity as well as on tumour growth and metastasis.
Methods
Quantitative phase imaging with digital holographic microscopy using HoloMonitor M4
The HoloMonitor M4 is a small time-lapse cytometer used for label-free live-cell imaging and quantitative analysis of monolayers of adhesive cells. The HoloMonitor M4 unit is equipped with a motorized xyz-stage, an Olympus PLN 20 × microscope objective, a light source in an external low-power laser unit (635 nm wavelength, 0.2 mW/cm2) and a 1.3 MP CMOS global shutter USB 2.0 camera. The technique employed, digital holographic microscopy, is based on measurements of phase shifts detected when a laser beam pass through living cells. The incoming laser is split into two beams, the sample beam and the reference beam. When the sample beam illuminates the cell, the light gets distorted when it passes through the cell, creating waves of light or phase shifts. When these waves are rejoined with the reference beam, an interference pattern, a hologram, is detected by the digital image sensor. Based on the hologram, a cell image or holographic image is reconstructed using a computer algorithm. This quantitative phase imaging method is used for analysis and quantification of various cellular features, like cell movement and parameters for cell morphology.Min kommentar
Jag hoppas ni gått in på den 14 sidor långa artikeln och inte missat sista stycket Supplementary information.
Där hittar ni 7 HoloMonitor videor som visar det texten beskriver.
Denna artikel behöver jag inte gissa på att den landar på PHI;s hemsida,förstasidan.
De norska forskarna har levererat ett marknadsföringsmaterial av högsta klass.
VD får skicka över den allra största chokladasken till Bergen University som ett litet tack för denna oförskämt braiga artikel,dessutom publicerad i en Nature edition.
Undertecknad ska å PHI,s aktieägarkollektivs vägnar be att få tacka våra norska vänner för det de åstadkommit. Greit. Hejja Norge.
Mvh the99
Har du för åsikter om detta?
SvaraRadera"Some limitations of the DHM system should be noted. As a cell population-based instrument with a 20 × objective, this DHM system has limitations regarding detection and quantification of fine protrusions as possible with other systems47. Nor is it possible to distinguish intracellular organelles (besides the nucleus to some degree), something which has been done with other QPI systems59"
Du är ihärdig och konsekvent,det vill jag komplimentera dig för :-D
RaderaMin åsikt är att det stärker författarnas trovärdighet.
De har letat efter tillkortakommanden hos HoloMonitor och är tydliga att lyfta fram den passagen i artikeln " Some limitations..." Alltså har de varit objektiva, och kritiska där det är befogat.Men det övriga innehållet (99%) är enligt mig ganska tydligt av vad de anser om instrumentet.
Jag bollar tillbaka en frågeställning till dig.
Vad har du för åsikt kring att GlycoImaging nu passerat det trängsta av de trånga nålsögon som kanske finnes (USA,s patentverk) och fått patentet godkänt?
Mvh the99
Det ju självklart jättebra. Frågan är om dom behöver söka FDA? Det blir väl ändå en form av diagnosverktyg Glycoimaging.
SvaraRaderaDock så tror jag projektet behöver köpas upp av en eller flera bolag som kan gasa på lite.